
Assessing Balance
Noah Greifer
2025-05-29
Source:vignettes/assessing-balance.Rmd
assessing-balance.RmdIntroduction
Covariate balance is the degree to which the distribution of covariates is similar across levels of the treatment. It has three main roles in causal effect estimation using matching: 1) as a target to optimize with matching, 2) as a method of assessing the quality of the resulting matches, and 3) as evidence to an audience that the estimated effect is close to the true effect. When covariate balance is achieved, the resulting effect estimate is less sensitive to model misspecification and ideally close to true treatment effect. The benefit of randomization is that covariate balance is achieved automatically (in expectation), which is why unadjusted effects estimated from randomized trial data (in the absence of drop-out) can be validly interpreted as causal effects. When using matching to recover causal effect estimates form observational data, balance is not guaranteed and must be assessed.
This document provides instructions for assessing and reporting
covariate balance as part of a matching analysis. The tools available in
MatchIt for balance assessment should be used during the
process of selecting a good matching scheme and ensuring that the chosen
scheme is adequate. These tools implement the recommendations of Ho et al. (2007) and
others for assessing balance.
In addition to the tools available in MatchIt, the
cobalt package has a suite of functions designed to assess
and display balance and is directly compatible with MatchIt
objects. cobalt has extensive documentation, but we
describe some of its functionality here as a complement to the tools in
MatchIt.
The structure of this document is as follows: first, we describe some
of the recommendations for balance checking and their rationale; next,
we describe the tools for assessing balance present in
MatchIt and display their use in evaluating several
matching schemes; finally; we briefly describe some of the functionality
in cobalt to extend that in MatchIt.
Recommendations for Balance Assessment
Assessing balance involves assessing whether the distributions of covariates are similar between the treated and control groups. Balance is typically assessed by examining univariate balance summary statistics for each covariate, though more complicated methods exist for assessing joint distributional balance as well. Visual depictions of distributional balance can be a helpful complement to numerical summaries, especially for hard to balance and prognostically important covariates.
Many recommendations for balance assessment have been described in the methodological literature. Unfortunately, there is no single best way to assess balance or to weigh balance summary statistics because the degree and form of balance that will yield the least bias in an effect estimate depends on unknown qualities of the outcome data-generating model. Nonetheless, there are a number of valuable recommendations that can be implemented to ensure matching is successful at eliminating or reducing bias. We review some of these here.
Common recommendations for assessing balance include the following:
Standardized mean differences. The standardized mean difference (SMD) is the difference in the means of each covariate between treatment groups standardized by a standardization factor so that it is on the same scale for all covariates. The standardization factor is typically the standard deviation of the covariate in the treated group when targeting the ATT or the pooled standard deviation across both groups when targeting the ATE. The standardization factor should be the same before and after matching to ensure changes in the mean difference are not confounded by changes in the standard deviation of the covariate. SMDs close to zero indicate good balance. Several recommended thresholds have been published in the literature; we recommend .1 and .05 for prognostically important covariates. Higher values may be acceptable when using covariate adjustment in the matched sample. In addition to computing SMDs on the covariates themselves, it is important to compute them on squares, cubes, and higher exponents as well as interactions between covariates. Several empirical studies have examined the appropriateness for using SMDs in balance assessment, including Belitser et al. (2011), Ali et al. (2014), and Stuart, Lee, and Leacy (2013); in general, there is often a high correlation between the mean or maximum absolute SMD and the degree of bias in the treatment effect.
Variance Ratios. The variance ratio is the ratio of the variance of a covariate in one group to that in the other. Variance ratios close to 1 indicate good balance because they imply the variances of the samples are similar (Austin 2009).
Empirical CDF Statistics. Statistics related to the difference in the empirical cumulative distribution functions (eCDFs) of each covariate between groups allow assessment of imbalance across the entire covariate distribution of that covariate rather than just its mean or variance. The maximum eCDF difference, also known as the Kolmogorov-Smirnov statistic, is sometimes recommended as a useful supplement to SMDs for assessing balance (Austin and Stuart 2015) and is often used as a criterion to use in propensity score methods that attempt to optimize balance (e.g., McCaffrey, Ridgeway, and Morral 2004; Diamond and Sekhon 2013). Although the mean eCDF difference has not been as well studied, it provides a summary of imbalance that may be missed by relying solely on the maximum difference.
Visual Diagnostics. Visual diagnostics such as eCDF plots, empirical quantile-quantile (eQQ) plots, and kernel density plots can be used to see exactly how the covariate distributions differ from each other, i.e., where in the distribution the greatest imbalances are (Ho et al. 2007; Austin 2009). This can help to figure out how to tailor a matching method to target imbalance in a specific region of the covariate distribution.
Prognostic scores. The prognostic score is an estimate of the potential outcome under control for each unit (Hansen 2008). Balance on the prognostic score has been shown to be highly correlated with bias in the effect estimate, making it a useful tool in balance assessment (Stuart, Lee, and Leacy 2013). Estimating the prognostic score requires having access to the outcome data, and using it may be seen as violating the principle of separating the design and analysis stages of a matching analysis (Rubin 2001). However, because only the outcome values from the control group are required to use the prognostic score, some separation is maintained.
Several multivariate statistics exist that summarize balance across the entire joint covariate distribution. These can be functions of the above measures, like the mean or maximum absolute SMD or the generalized weighted distance [GWD; Franklin et al. (2014)], which is the sum of SMDs for the covariates and their squares and interactions, or separate statistics that measure quantities that abstract away from the distribution of individual covariates, like the L1 distance (Iacus, King, and Porro 2011), cross-match test (Heller, Rosenbaum, and Small 2010), or energy distance (Huling and Mak 2020).
Balance on the propensity score has often been considered a useful measure of balance, but we do not necessarily recommend it except as a supplement to balance on the covariates. Propensity score balance will generally be good with any matching method regardless of the covariate balancing potential of the propensity score, so a balanced propensity score does not imply balanced covariates (Austin 2009). Similarly, it may happen that covariates may be well balanced even if the propensity score is not balanced, such as when covariates are prioritized above the propensity score in the matching specification (e.g., with genetic matching). Given these observations, the propensity score should not be relied upon for assessing covariate balance. Simulation studies by Stuart, Lee, and Leacy (2013) provide evidence for this recommendation against relying on propensity score balance.
There has been some debate about the use of hypothesis tests, such as
t-tests or Kolmogorov-Smirnov tests, for assessing covariate balance.
The idea is that balance tests test the null hypothesis that the matched
sample has equivalent balance to a randomized experiment. There are
several problems with balance tests, described by Ho et al. (2007) and
Imai, King, and Stuart (2008): 1) balance is a property of the
sample, not a of a population from which the sample was drawn; 2) the
power of balance tests depends on the sample size, which changes during
matching even if balance does not change; and 3) the use of hypothesis
tests implies a uniform decision criterion for rejecting the null
hypothesis (e.g., p-value less than .05, potentially with corrections
for multiple comparisons), when balance should be improved without
limit. MatchIt does not report any balance tests or
p-values, instead relying on the descriptive statistics described
above.
Recommendations for Balance Reporting
A variety of methods should be used when assessing balance to try to find an optimal matched set that will ideally yield a low-error estimate of the desired effect. However, reporting every balance statistic or plot in a research report or publication can be burdensome and unnecessary. That said, it is critical to report balance to demonstrate to readers that the resulting estimate is approximately unbiased and relies little on extrapolation or correct outcome model specification. We recommend the following in reporting balance in a matching analysis:
Report SMDs before and after matching for each covariate, any prognostically important interactions between covariates, and the prognostic score; this can be reported in a table or in a Love plot.
Report summaries of balance for other statistics, e.g., the largest mean and maximum eCDF difference among the covariates and the largest SMD among squares, cubes, and interactions of the covariates.
MatchIt provides tools for calculating each of these
statistics so they can be reported with ease in a manuscript or
report.
Assessing Balance with MatchIt
MatchIt contains several tools to assess balance
numerically and graphically. The primary balance assessment function is
summary.matchit(), which is called when using
summary() on a MatchIt object and produces
several tables of balance statistics before and after matching.
plot.summary.matchit() generates a Love plot using R’s base
graphics system containing the standardized mean differences resulting
from a call to summary.matchit() and provides a nice way to
display balance visually for inclusion in an article or report.
plot.matchit() generates several plots that display
different elements of covariate balance, including propensity score
overlap and distribution plots of the covariates. These functions
together form a suite that can be used to assess and report balance in a
variety of ways.
To demonstrate MatchIt’s balance assessment
capabilities, we will use the Lalonde data included in
MatchIt and used in vignette("MatchIt"). We
will perform 1:1 nearest neighbor matching with replacement on the
propensity score, though the functionality is identical across all
matching methods except propensity score subclassification, which we
illustrate at the end.
library("MatchIt")
data("lalonde", package = "MatchIt")
#1:1 NN matching w/ replacement on a logistic regression PS
m.out <- matchit(treat ~ age + educ + race + married +
nodegree + re74 + re75, data = lalonde,
replace = TRUE)
m.out## A `matchit` object
## - method: 1:1 nearest neighbor matching with replacement
## - distance: Propensity score
## - estimated with logistic regression
## - number of obs.: 614 (original), 267 (matched)
## - target estimand: ATT
## - covariates: age, educ, race, married, nodegree, re74, re75
summary.matchit()
When summary() is called on a matchit
object, several tables of information are displayed. These include
balance statistics for each covariate before matching, balance
statistics for each covariate after matching, the percent reduction in
imbalance after matching, and the sample sizes before and after
matching. summary.matchit() has four additional arguments
that control how balance is computed:
-
interactionscontrols whether balance statistics for all squares and pairwise interactions of covariates are to be displayed in addition to the covariates. The default isFALSE, and setting toTRUEcan make the output massive when many covariates are present, but it is important to ensure no important interactions remain imbalanced. -
addlvariablesallows for balance to be assessed on variables other than those inside thematchitobject. For example, if the distance between units only relied on a subset of covariates but balance needed to be achieved on all covariates,addlvariablescould be used to supply these additional covariates. In addition to adding other variables,addlvariablescan be used to request balance on specific functions of the covariates already in thematchitobject, such as polynomial terms or interactions. The input toaddlvariablescan be a one-sided formula with the covariates and any desired transformations thereof on the right hand side, just like a model formula (e.g.,addlvariables = ~ X1 + X2 + I(X1^2)would request balance onX1,X2, and the square ofX1). Additional variables supplied toaddlvariablesbut not present in thematchitobject can be supplied as a data frame using thedataargument. -
standardizecontrols whether standardized or unstandardized statistics are to displayed. Standardized statistics include the standardized mean difference and eCDF statistics; unstandardized statistics include the raw difference in means and eQQ plot statistics. (Regardless, the variance ratio will always be displayed.). The default isTRUEfor standardized statistics, which are more common to report because they are all on the same scale regardless of the scale of the covariates1. -
pair.distcontrols whether within-pair distances should be computed and displayed. These reflect the average distance between units within the same pair, standardized or unstandardized according to the argument tostandardize. The default isTRUE. With full matching, exact matching, coarsened exact matching, and propensity score subclassification, computing pair distances can take a long time, and so it may be beneficial to set toFALSEin these cases.
In addition, the arguments un (default:
TRUE) and improvement (default:
FALSE) control whether balance prior to matching should be
displayed and whether the percent balance improvement after matching
should be displayed. These can be set to FALSE to reduce
the output.
Below, we call summary.matchit() with
addlvariables to display balance on covariates and a few
functions of them in the matched sample. In particular, we request
balance on the square of age, the variables representing
whether re74 and re75 were equal to 0, and the
interaction between educ and race.
##
## Call:
## matchit(formula = treat ~ age + educ + race + married + nodegree +
## re74 + re75, data = lalonde, replace = TRUE)
##
## Summary of Balance for All Data:
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max
## distance 0.577 0.182 1.794 0.921 0.377 0.644
## age 25.816 28.030 -0.309 0.440 0.081 0.158
## educ 10.346 10.235 0.055 0.496 0.035 0.111
## raceblack 0.843 0.203 1.762 . 0.640 0.640
## racehispan 0.059 0.142 -0.350 . 0.083 0.083
## racewhite 0.097 0.655 -1.882 . 0.558 0.558
## married 0.189 0.513 -0.826 . 0.324 0.324
## nodegree 0.708 0.597 0.245 . 0.111 0.111
## re74 2095.574 5619.237 -0.721 0.518 0.225 0.447
## re75 1532.055 2466.484 -0.290 0.956 0.134 0.288
## I(age^2) 717.395 901.779 -0.428 0.363 0.081 0.158
## I(re74 == 0)TRUE 0.708 0.261 0.983 . 0.447 0.447
## I(re75 == 0)TRUE 0.600 0.312 0.587 . 0.288 0.288
## educ:raceblack 8.697 2.047 1.580 0.980 0.354 0.645
## educ:racehispan 0.578 1.263 -0.294 0.487 0.046 0.078
## educ:racewhite 1.070 6.925 -1.767 0.365 0.279 0.555
##
## Summary of Balance for Matched Data:
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max Std. Pair Dist.
## distance 0.577 0.576 0.004 0.992 0.003 0.049 0.013
## age 25.816 24.103 0.239 0.557 0.077 0.341 1.262
## educ 10.346 10.378 -0.016 0.577 0.022 0.059 1.086
## raceblack 0.843 0.838 0.015 . 0.005 0.005 0.045
## racehispan 0.059 0.065 -0.023 . 0.005 0.005 0.297
## racewhite 0.097 0.097 0.000 . 0.000 0.000 0.054
## married 0.189 0.130 0.152 . 0.059 0.059 0.511
## nodegree 0.708 0.703 0.012 . 0.005 0.005 0.868
## re74 2095.574 2336.463 -0.049 1.036 0.041 0.216 0.609
## re75 1532.055 1503.929 0.009 2.129 0.068 0.238 0.650
## I(age^2) 717.395 670.946 0.108 0.510 0.077 0.341 1.196
## I(re74 == 0)TRUE 0.708 0.492 0.476 . 0.216 0.216 0.975
## I(re75 == 0)TRUE 0.600 0.362 0.485 . 0.238 0.238 1.037
## educ:raceblack 8.697 8.589 0.026 0.869 0.024 0.054 0.468
## educ:racehispan 0.578 0.638 -0.026 0.827 0.007 0.022 0.336
## educ:racewhite 1.070 1.151 -0.024 0.846 0.005 0.022 0.220
##
## Sample Sizes:
## Control Treated
## All 429. 185
## Matched (ESS) 46.31 185
## Matched 82. 185
## Unmatched 347. 0
## Discarded 0. 0Let’s examine the output in detail. The first table
(Summary of Balance for All Data) provides balance in the
sample prior to matching. The included statistics are the mean of the
covariates in the treated group (Means Treated), the mean
of the covariate in the control group (Means Control), the
SMDs (Std. Mean Diff.), the variance ratio
(Var. Ratio), the average distance between the eCDFs of the
covariate across the groups (eCDF Mean), and the largest
distance between the eCDFs (eCDF Max). Setting
un = FALSE would have suppressed the creation of this
table.
The second table (Summary of Balance for Matched Data)
contains all the same statistics in the matched sample. Because we
implicitly request pair distance, an additional column for standardized
pair distances (Std. Pair Dist.) is displayed.
The final table (Sample Sizes) contains the sizes of the
samples before (All) and after (Matched)
matching, as well as the number of units left unmatched
(Unmatched) and the number of units dropped due to a common
support restriction (Discarded).
The SMDs are computed as the mean difference divided by a standardization factor computed in the unmatched sample. An absolute SMD close to 0 indicates good balance; although a number of recommendations for acceptable values have appeared in the literature, we recommend absolute values less than .1 and less than .05 for potentially prognostically important variables.
The variance ratios are computed as the ratio of the variance of the treated group to that of the control group for each covariate. Variance ratios are not computed for binary covariates because they are a function of the prevalence in each group, which is captured in the mean difference and eCDF statistics. A variance ratio close to 1 indicates good balance; a commonly used recommendation is for variance ratios to be between .5 and 2.
The eCDF statistics correspond to the difference in the overall
distributions of the covariates between the treatment groups. The values
of both statistics range from 0 to 1, with values closer to zero
indicating better balance. There are no specific recommendations for the
values these statistics should take, though notably high values may
indicate imbalance on higher moments of the covariates. The eQQ
statistics produced when standardize = FALSE are
interpreted similarly but are on the scale of the covariate.
All these statistics should be considered together. Imbalance as measured by any of them may indicate a potential failure of the matching scheme to achieve distributional balance.
plot.summary.matchit()
A Love plot is a clean way to visually summarize balance. Using
plot on the output of a call to summary() on a
matchit object produces a Love plot of the standardized
mean differences. plot.summary.matchit() has several
additional arguments that can be used to customize the plot.
-
abscontrols whether standardized mean difference should be displayed in absolute value or not. Default isTRUE. -
var.ordercontrols how the variables are ordered on the y-axis. The options are"data"(the default), which orders the variables as they appear the in thesummary.matchit()output;"unmatched", which orders the variables based on their standardized mean differences before matching;"matched", which orders the variables based on their standardized mean differences after matching; and"alphabetical", which orders the variables alphabetically. Using"unmatched"tends to result in attractive plots and ensures the legend doesn’t overlap with points in its default position. -
thresholdcontrols where vertical lines indicating chosen thresholds should appear on the x-axis. Should be a numeric vector. The default isc(.1, .05), which display vertical lines at .1 and .05 standardized mean difference units. -
positioncontrols the position of the legend. The default is"bottomright", which puts the legend in the bottom right corner of the plot, and any keyword value available to supplied toxinlegend()is allowed.
Below we create a Love plot of the covariates.
m.sum <- summary(m.out, addlvariables = ~ I(age^2) + I(re74==0) +
I(re75==0) + educ:race)
plot(m.sum, var.order = "unmatched")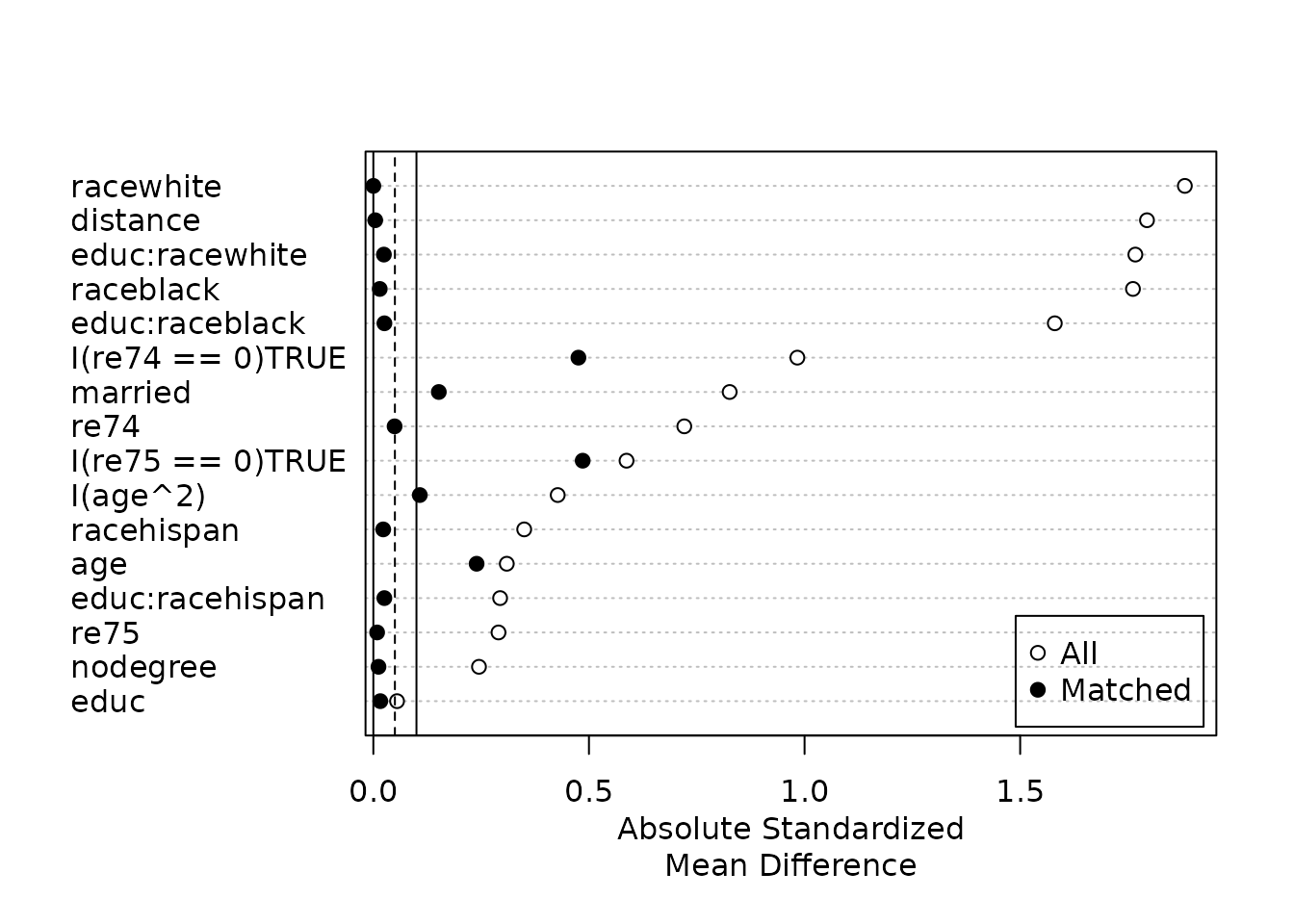
From this plot it is clear to see that balance was quite poor prior
to matching, but full matching improved balance on all covariates, and
most within a threshold of .1. To make the variable names cleaner, the
original variables should be renamed prior to matching.
cobalt provides many additional options to generate and
customize Love plots using the love.plot() function and
should be used if a plot beyond what is available with
plot.summary.matchit() is desired.
plot.matchit()
In addition to numeric summaries of balance, MatchIt
offers graphical summaries as well using plot.matchit()
(i.e., using plot() on a matchit object). We
can create eQQ plots, eCDF plots, or density plots of the covariates and
histograms or jitter plots of the propensity score. The covariate plots
can provide a summary of the balance of the full marginal distribution
of a covariate beyond just the mean and variance.
plot.matchit() has a few arguments to customize the
output:
-
typecorresponds to the type of plot desired. Options include"qq"for eQQ plots (the default),"ecdf"for eCDF plots,"density"for density plots (or bar plots for categorical variables),"jitter"for jitter plots, and"histogram"for histograms. -
interactivecontrols whether the plot is interactive or not. For eQQ, eCDF, and density plots, this allows us to control when the next page of covariates is to be displayed since only three can appear at a time. For jitter plots, this can allow us to select individual units with extreme values for further inspection. The default isTRUE. -
which.xsis used to specify for which covariates to display balance in eQQ, eCDF, and density plots. The default is to display balance on all, but we can request balance just on a specific subset. If three or fewer are requested,interactiveis ignored. The argument can be supplied as a one-sided formula with the variables of interest on the right or a character vector containing the names of the desired variables. If any variables are not in thematchitobject, adataargument can be supplied with a data set containing the named variables.
Below, we demonstrate the eQQ plot:
#eQQ plot
plot(m.out, type = "qq", which.xs = ~age + nodegree + re74)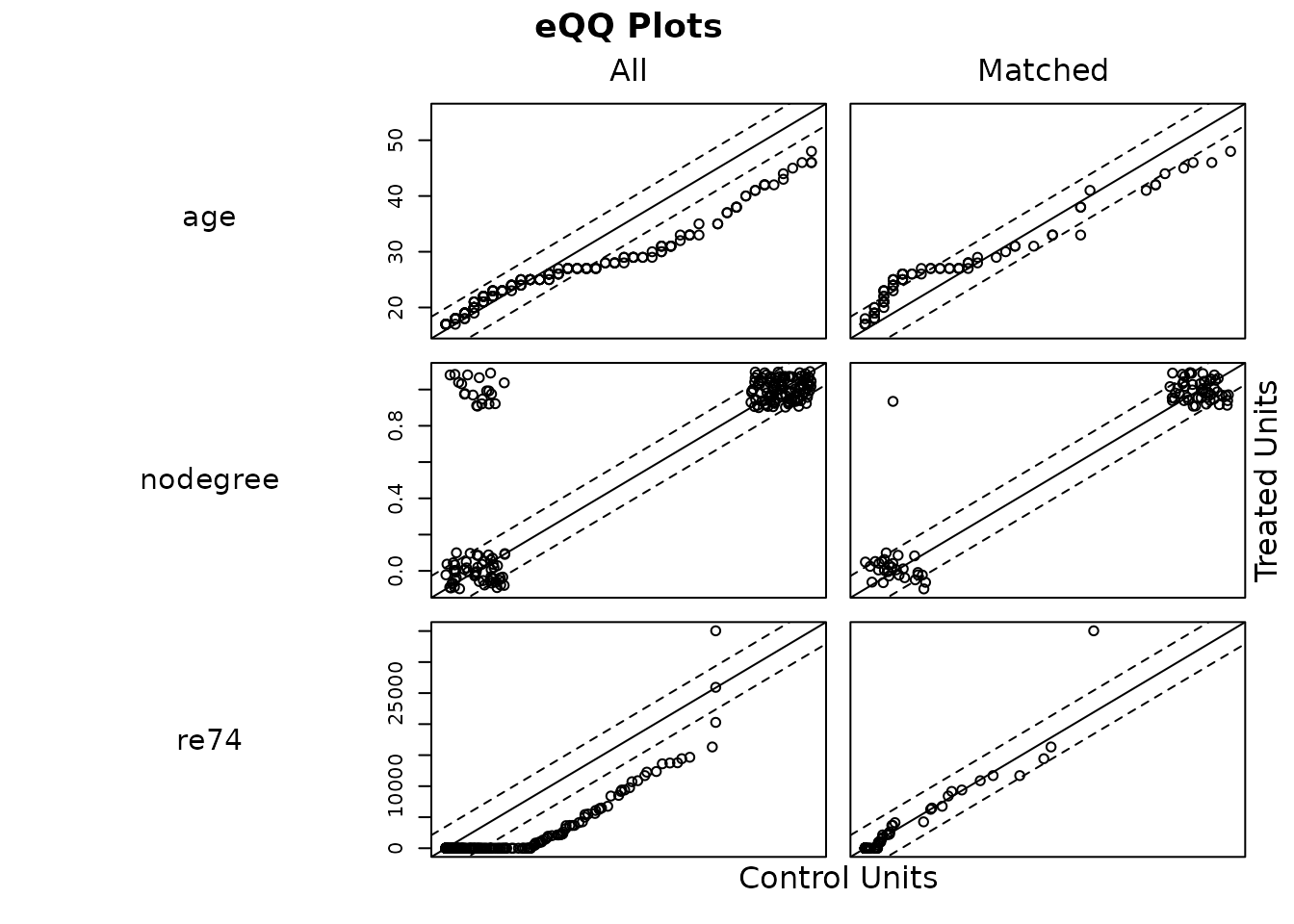
The y-axis displays the each value of the covariate for the treated
units, and the x-axis displays the the value of the covariate at the
corresponding quantile in the control group. When values fall on the 45
degree line, the groups are balanced. Above, we can see that
age remains somewhat imbalanced, but nodegree
and re74 have much better balance after matching than
before. The difference between the x and y values of each point are used
to compute the eQQ difference statistics that are displayed in
summary.matchit() with
standardize = FALSE.
Below, we demonstrate the eCDF plot:
#eCDF plot
plot(m.out, type = "ecdf", which.xs = ~educ + married + re75)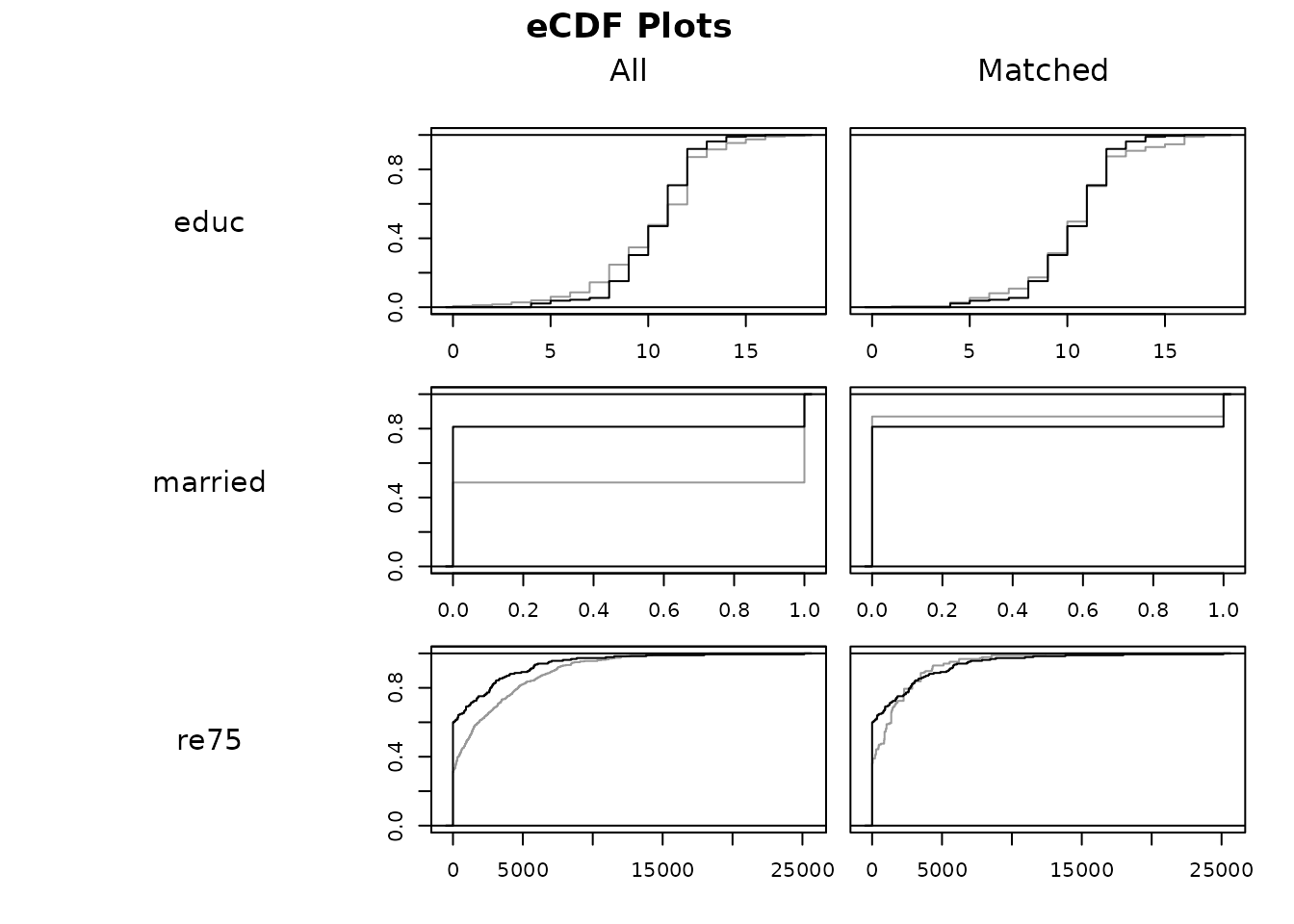
The x-axis displays the covariate values and the y-axis displays the
proportion of the sample at or less than that covariate value. Perfectly
overlapping lines indicate good balance. The black line corresponds to
the treated group and the gray line to the control group. Although
educ and re75 were fairly well balanced before
matching, their balance has improved nonetheless. married
appears far better balanced after matching than before. The vertical
difference between the eCDFs lines of each treatment group is used to
compute the eCDF difference statistics that are displayed in
summary.matchit() with standardize = TRUE.
Below, we demonstrate the density plot:
#density plot
plot(m.out, type = "density", which.xs = ~age + educ + race)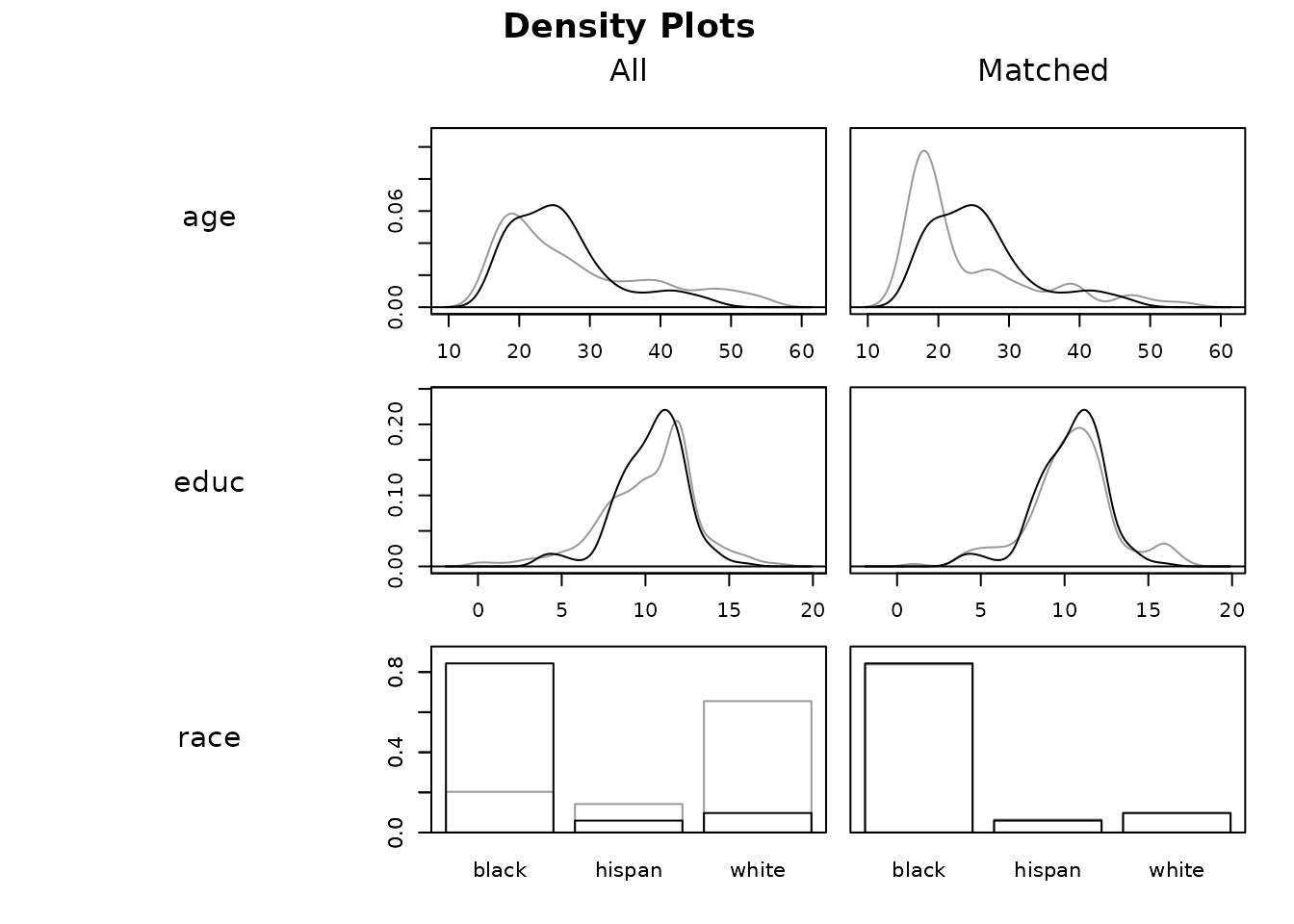
The x-axis displays the covariate values and the y-axis displays the density of the sample at that covariate value. For categorical variables, the y-axis displays the proportion of the sample at that covariate value. The black line corresponds to the treated group and the gray line to the control group. Perfectly overlapping lines indicate good balance. Density plots display similar information to eCDF plots but may be more intuitive for some users because of their link to histograms.
Assessing Balance After Subclassification
With subclassification, balance can be checked both within each
subclass and overall. With summary.matchit(), we can
request to view balance only in aggregate or in each subclass. The
latter can help us decide if we can interpret effects estimated within
each subclass as unbiased. The plot.summary.matchit() and
plot.matchit() outputs can be requested either in aggregate
or for each subclass. We demonstrate this below. First we will perform
propensity score subclassification using 4 subclasses (typically more is
beneficial).
#Subclassification on a logistic regression PS
s.out <- matchit(treat ~ age + educ + race + married +
nodegree + re74 + re75, data = lalonde,
method = "subclass", subclass = 4)
s.out## A `matchit` object
## - method: Subclassification (4 subclasses)
## - distance: Propensity score
## - estimated with logistic regression
## - number of obs.: 614 (original), 614 (matched)
## - target estimand: ATT
## - covariates: age, educ, race, married, nodegree, re74, re75When using summary(), the default is to display balance
only in aggregate using the subclassification weights. This balance
output looks similar to that for other matching methods.
summary(s.out)##
## Call:
## matchit(formula = treat ~ age + educ + race + married + nodegree +
## re74 + re75, data = lalonde, method = "subclass", subclass = 4)
##
## Summary of Balance for All Data:
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max
## distance 0.577 0.182 1.794 0.921 0.377 0.644
## age 25.816 28.030 -0.309 0.440 0.081 0.158
## educ 10.346 10.235 0.055 0.496 0.035 0.111
## raceblack 0.843 0.203 1.762 . 0.640 0.640
## racehispan 0.059 0.142 -0.350 . 0.083 0.083
## racewhite 0.097 0.655 -1.882 . 0.558 0.558
## married 0.189 0.513 -0.826 . 0.324 0.324
## nodegree 0.708 0.597 0.245 . 0.111 0.111
## re74 2095.574 5619.237 -0.721 0.518 0.225 0.447
## re75 1532.055 2466.484 -0.290 0.956 0.134 0.288
##
## Summary of Balance Across Subclasses
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max
## distance 0.577 0.539 0.173 0.678 0.062 0.126
## age 25.816 24.975 0.118 0.465 0.085 0.297
## educ 10.346 10.433 -0.043 0.582 0.023 0.061
## raceblack 0.843 0.767 0.210 . 0.076 0.076
## racehispan 0.059 0.042 0.076 . 0.018 0.018
## racewhite 0.097 0.191 -0.318 . 0.094 0.094
## married 0.189 0.196 -0.017 . 0.007 0.007
## nodegree 0.708 0.657 0.113 . 0.051 0.051
## re74 2095.574 2557.709 -0.095 0.968 0.048 0.264
## re75 1532.055 1490.040 0.013 1.505 0.035 0.146
##
## Sample Sizes:
## Control Treated
## All 429. 185
## Matched (ESS) 102.3 185
## Matched 429. 185
## Unmatched 0. 0
## Discarded 0. 0An additional option in summary(),
subclass, allows us to request balance for individual
subclasses. subclass can be set to TRUE to
display balance for all subclasses or the indices of individual
subclasses for which balance is to be displayed. Below we call
summary() and request balance to be displayed on all
subclasses (setting un = FALSE to suppress balance in the
original sample):
summary(s.out, subclass = TRUE, un = FALSE)##
## Call:
## matchit(formula = treat ~ age + educ + race + married + nodegree +
## re74 + re75, data = lalonde, method = "subclass", subclass = 4)
##
## Summary of Balance by Subclass:
##
## - Subclass 1
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max
## distance 0.239 0.095 0.879 2.494 0.313 0.508
## age 26.478 28.800 -0.345 0.394 0.090 0.160
## educ 10.304 10.214 0.040 0.620 0.025 0.084
## raceblack 0.370 0.063 0.635 . 0.307 0.307
## racehispan 0.239 0.167 0.169 . 0.072 0.072
## racewhite 0.391 0.770 -0.776 . 0.379 0.379
## married 0.370 0.589 -0.455 . 0.219 0.219
## nodegree 0.587 0.584 0.007 . 0.003 0.003
## re74 5430.539 6363.913 -0.118 1.298 0.087 0.284
## re75 2929.039 2699.399 0.054 1.587 0.047 0.144
##
## - Subclass 2
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max
## distance 0.604 0.612 -0.214 0.905 0.083 0.195
## age 25.556 24.409 0.152 0.461 0.114 0.370
## educ 9.933 9.773 0.066 0.448 0.084 0.188
## raceblack 1.000 1.000 0.000 . 0.000 0.000
## racehispan 0.000 0.000 0.000 . 0.000 0.000
## racewhite 0.000 0.000 0.000 . 0.000 0.000
## married 0.378 0.091 0.592 . 0.287 0.287
## nodegree 0.667 0.500 0.354 . 0.167 0.167
## re74 1777.422 2516.589 -0.219 0.433 0.076 0.280
## re75 972.344 1131.077 -0.100 0.666 0.034 0.086
##
## - Subclass 3
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max
## distance 0.693 0.691 0.130 1.281 0.055 0.189
## age 24.021 22.964 0.158 0.509 0.128 0.281
## educ 10.170 10.286 -0.069 1.040 0.038 0.099
## raceblack 1.000 1.000 0.000 . 0.000 0.000
## racehispan 0.000 0.000 0.000 . 0.000 0.000
## racewhite 0.000 0.000 0.000 . 0.000 0.000
## married 0.021 0.107 -0.595 . 0.086 0.086
## nodegree 0.681 0.750 -0.148 . 0.069 0.069
## re74 939.969 888.947 0.020 2.038 0.059 0.216
## re75 1217.455 1285.387 -0.018 1.535 0.038 0.188
##
## - Subclass 4
## Means Treated Means Control Std. Mean Diff. Var. Ratio eCDF Mean eCDF Max
## distance 0.767 0.753 0.540 2.961 0.165 0.445
## age 27.213 23.786 0.461 0.521 0.150 0.459
## educ 10.957 11.429 -0.341 0.701 0.059 0.126
## raceblack 1.000 1.000 0.000 . 0.000 0.000
## racehispan 0.000 0.000 0.000 . 0.000 0.000
## racewhite 0.000 0.000 0.000 . 0.000 0.000
## married 0.000 0.000 0.000 . 0.000 0.000
## nodegree 0.894 0.786 0.350 . 0.108 0.108
## re74 291.783 540.618 -0.307 0.917 0.083 0.280
## re75 1015.289 854.751 0.079 3.523 0.112 0.266
##
## Sample Sizes by Subclass:
## 1 2 3 4 All
## Control 365 22 28 14 429
## Treated 46 45 47 47 185
## Total 411 67 75 61 614We can plot the standardized mean differences in a Love plot that
also displays balance for the subclasses using
plot.summary.matchit() on a summary.matchit()
object with subclass = TRUE.
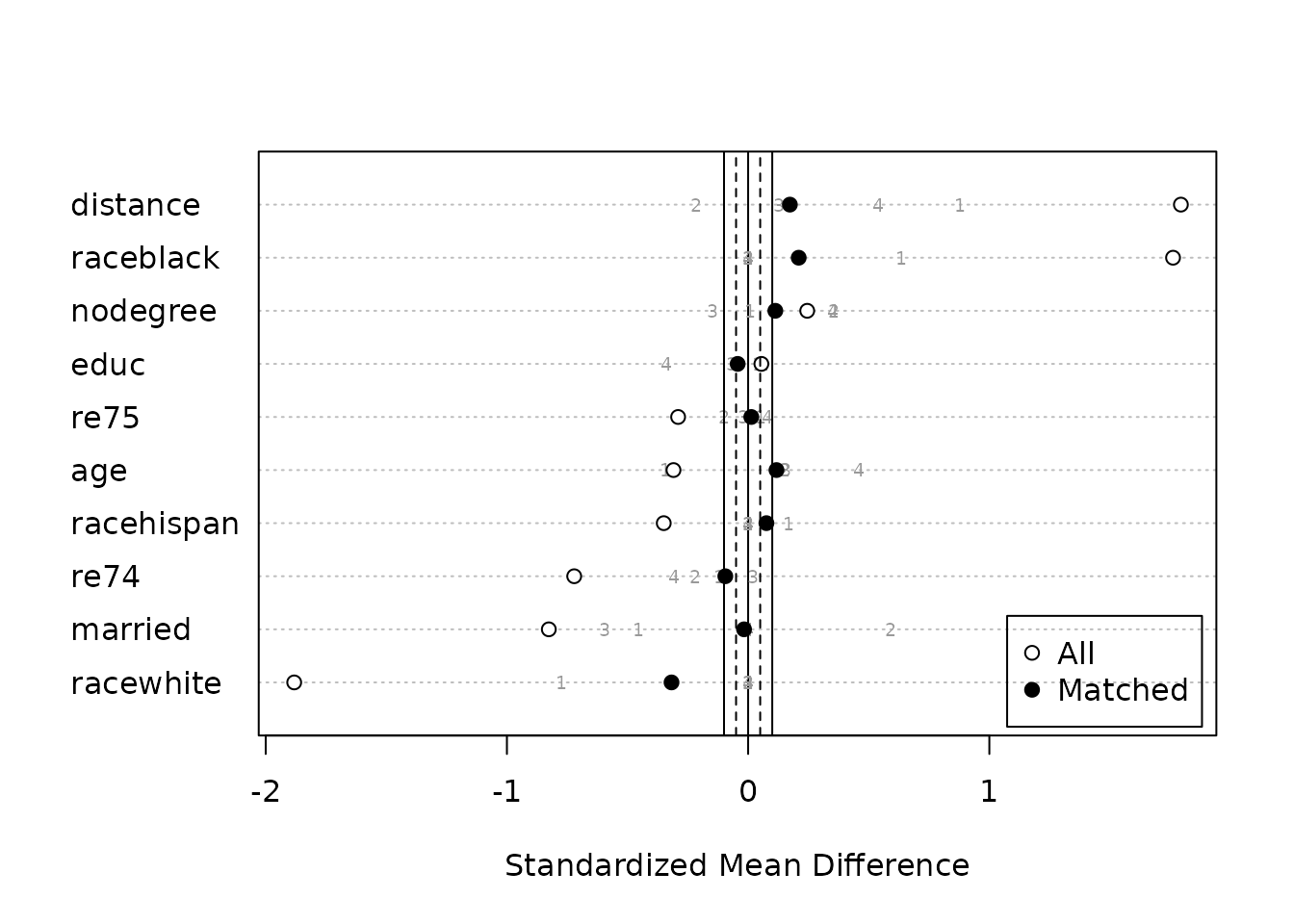
Note that for some variables, while the groups are balanced in aggregate (black dots), the individual subclasses (gray numbers) may not be balanced, in which case unadjusted effect estimates within these subclasses should not be interpreted as unbiased.
When we plot distributional balance using
plot.matchit(), again we can choose whether balance should
be displayed in aggregate or within subclasses again using the
subclass option, which functions the same as it does with
summary.matchit(). Below we demonstrate checking balance
within a subclass.
plot(s.out, type = "density", which.xs = ~educ + married + re75,
subclass = 1)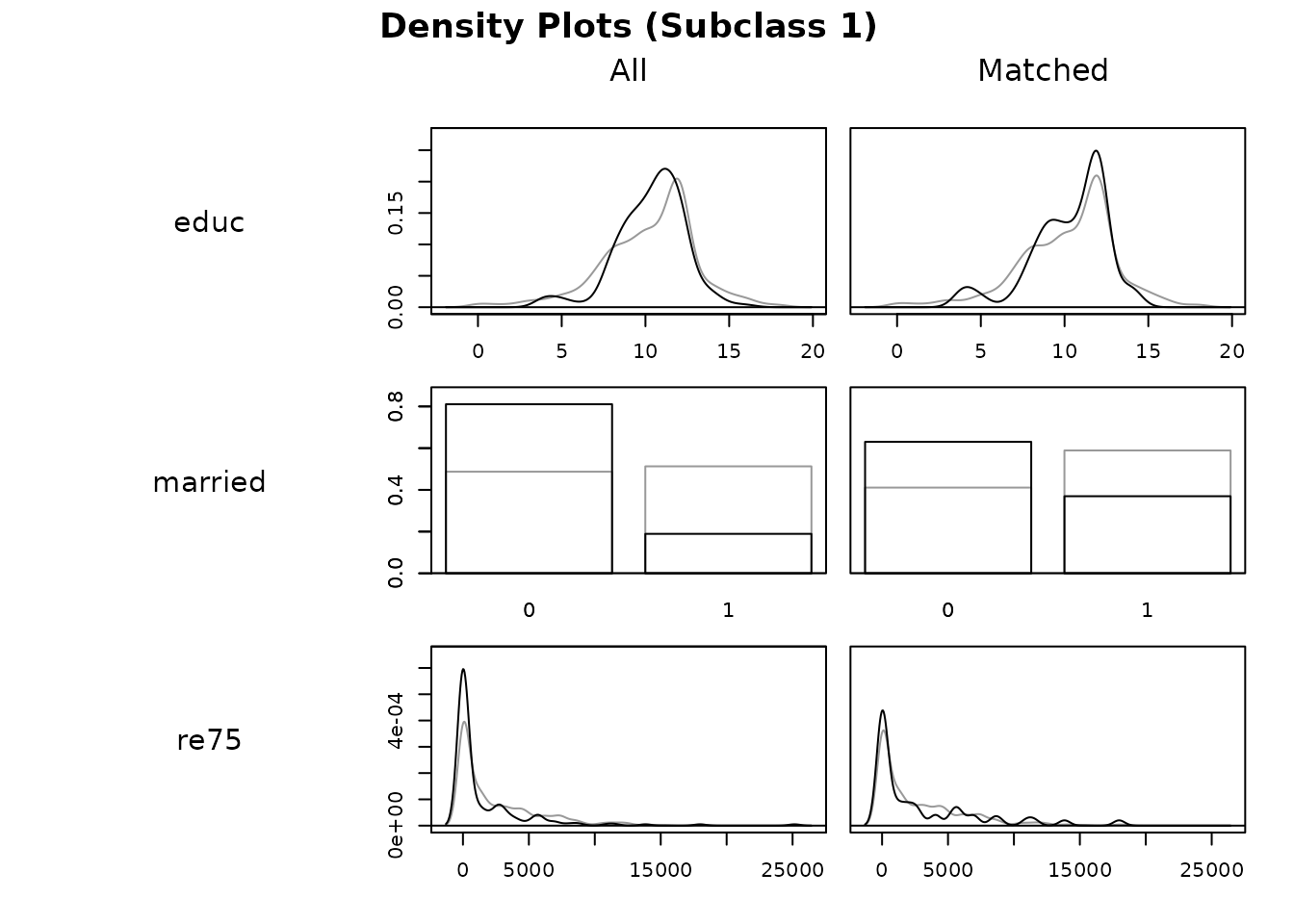
If we had set subclass = FALSE, plots would have been
displayed in aggregate using the subclassification weights. If
subclass is unspecified, a prompt will ask us for which
subclass we want to see balance.
Assessing Balance with cobalt
The cobalt package was designed specifically for
checking balance before and after matching (and weighting). It offers
three main functions, bal.tab(), love.plot(),
and bal.plot(), which perform similar actions to
summary.matchit(), plot.summary.matchit(), and
plot.matchit(), respectively. These functions directly
interface with matchit objects, making cobalt
straightforward to use in conjunction with MatchIt.
cobalt can be used as a complement to MatchIt,
especially for more advanced uses that are not accommodated by
MatchIt, such as comparing balance across different
matching schemes and even different packages, assessing balance in
clustered or multiply imputed data, and assessing balance with
multi-category, continuous, and time-varying treatments. The main
cobalt vignette
(vignette("cobalt", package = "cobalt")) contains many
examples of its use with MatchIt objects, so we only
provide a short demonstration of its capabilities here.
bal.tab()
bal.tab() produces tables of balance statistics similar
to summary.matchit(). The columns displayed can be
customized to limit how much information is displayed and isolate
desired information. We call bal.tab() with a few of its
options specified below:
## Balance Measures
## Type Diff.Un V.Ratio.Un KS.Un Diff.Adj V.Ratio.Adj KS.Adj
## distance Distance 1.794 0.921 0.644 0.004 0.992 0.049
## age Contin. -0.309 0.440 0.158 0.239 0.557 0.341
## educ Contin. 0.055 0.496 0.111 -0.016 0.577 0.059
## race_black Binary 0.640 . 0.640 0.005 . 0.005
## race_hispan Binary -0.083 . 0.083 -0.005 . 0.005
## race_white Binary -0.558 . 0.558 0.000 . 0.000
## married Binary -0.324 . 0.324 0.059 . 0.059
## nodegree Binary 0.111 . 0.111 0.005 . 0.005
## re74 Contin. -0.721 0.518 0.447 -0.049 1.036 0.216
## re75 Contin. -0.290 0.956 0.288 0.009 2.129 0.238
##
## Sample sizes
## Control Treated
## All 429. 185
## Matched (ESS) 46.31 185
## Matched (Unweighted) 82. 185
## Unmatched 347. 0The output is very similar to that of summary.matchit(),
except that the balance statistics computed before matching (with the
suffix .Un) and those computed after matching (with the
suffix .Adj) are in the same table. By default, only SMDs
after matching (Diff.Adj) are displayed; by setting
un = TRUE, we requested that the balance statistics before
matching also be displayed, and by setting
stats = c("m", "v", "ks") we requested mean differences,
variance ratios, and Kolmogorov-Smirnov statistics. Other balance
statistics and summary statistics can be requested as well. One
important detail to note is that the default for binary covariates is to
print the raw difference in proportion rather than the standardized mean
difference, so there will be an apparent discrepancy for these variables
between bal.tab() and summary.matchit()
output, though this behavior can be changed by setting
binary = "std" in the call to bal.tab().
Functionality for producing balance statistics for additional variables
and for powers and interactions of the covariates is available using the
addl, poly, and int options.
bal.tab() and other cobalt functions can
produce balance not just on a single matchit object but on
several at the same time, which facilitates comparing balance across
several matching specifications. For example, if we wanted to compare
the full matching results to the results of nearest neighbor matching
without replacement, we could supply both to bal.tab(),
which we demonstrate below:
#Nearest neighbor (NN) matching on the PS
m.out2 <- matchit(treat ~ age + educ + race + married +
nodegree + re74 + re75, data = lalonde)
#Balance on covariates after full and NN matching
bal.tab(treat ~ age + educ + race + married +
nodegree + re74 + re75, data = lalonde,
un = TRUE, weights = list(full = m.out, nn = m.out2))## Balance Measures
## Type Diff.Un Diff.full Diff.nn
## age Contin. -0.309 0.239 0.072
## educ Contin. 0.055 -0.016 -0.129
## race_black Binary 0.640 0.005 0.373
## race_hispan Binary -0.083 -0.005 -0.157
## race_white Binary -0.558 0.000 -0.216
## married Binary -0.324 0.059 -0.022
## nodegree Binary 0.111 0.005 0.070
## re74 Contin. -0.721 -0.049 -0.050
## re75 Contin. -0.290 0.009 -0.026
##
## Effective sample sizes
## Control Treated
## All 429. 185
## full 46.31 185
## nn 185. 185This time, we supplied bal.tab() with the covariates and
dataset and supplied the matchit output objects in the
weights argument (which extracts the matching weights from
the objects). Here we can see that full matching yields better balance
than nearest neighbor matching overall, though balance is slightly worse
for age and maried and the effective sample
size is lower.
love.plot()
love.plot() creates a Love plot of chosen balance
statistics. It offers many options for customization, including the
shape and colors of the points, how the variable names are displayed,
and for which statistics balance is to be displayed. Below is an example
of its basic use:
love.plot(m.out, binary = "std")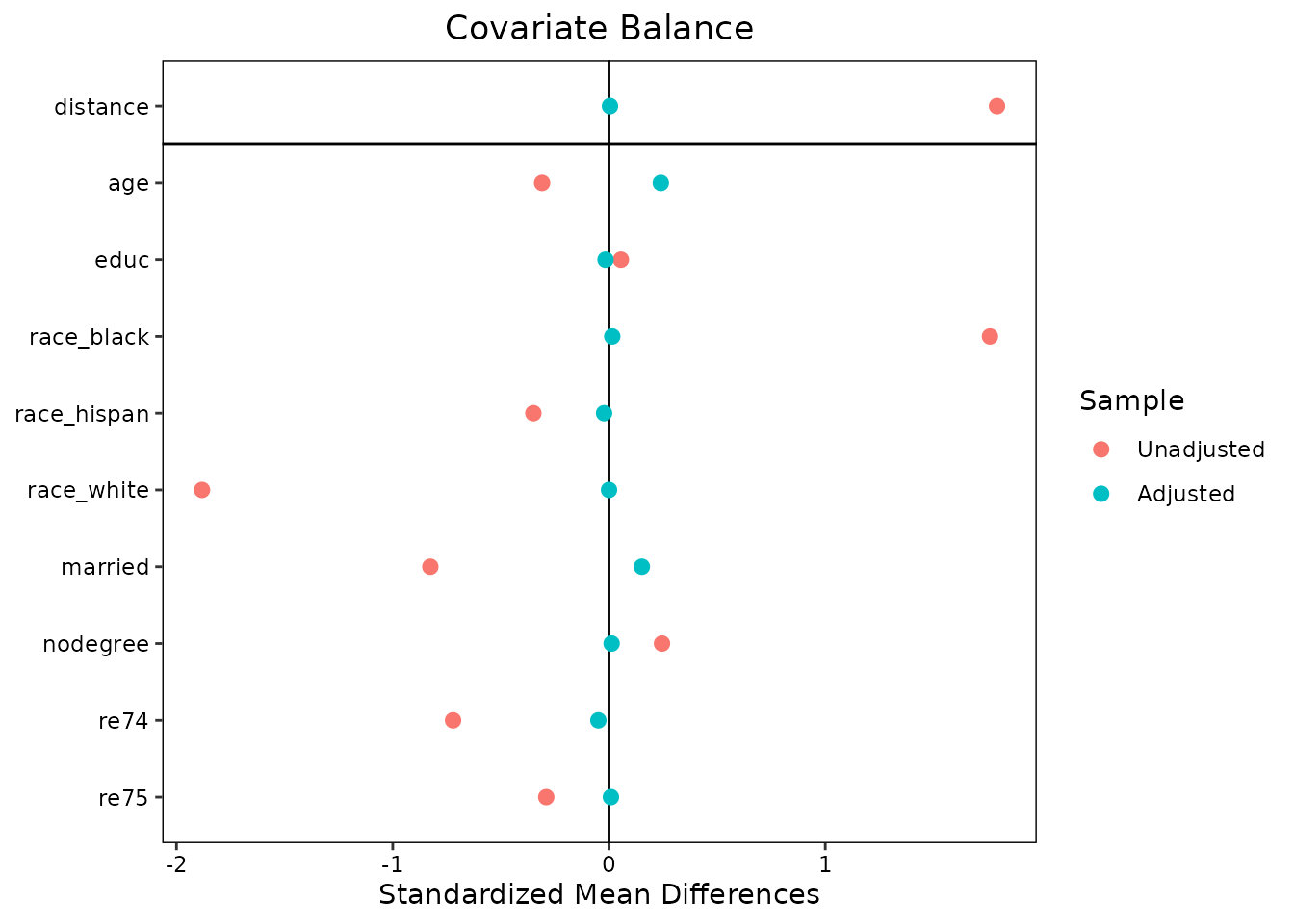
The syntax is straightforward and similar to that of
bal.tab(). Below we demonstrate a more advanced use that
customizes the appearance of the plot and displays balance not only on
mean differences but also on Kolmogorov-Smirnov statistics and for both
full matching and nearest neighbor matching simultaneously.
love.plot(m.out, stats = c("m", "ks"), poly = 2, abs = TRUE,
weights = list(nn = m.out2),
drop.distance = TRUE, thresholds = c(m = .1),
var.order = "unadjusted", binary = "std",
shapes = c("circle filled", "triangle", "square"),
colors = c("red", "blue", "darkgreen"),
sample.names = c("Original", "Full Matching", "NN Matching"),
position = "bottom")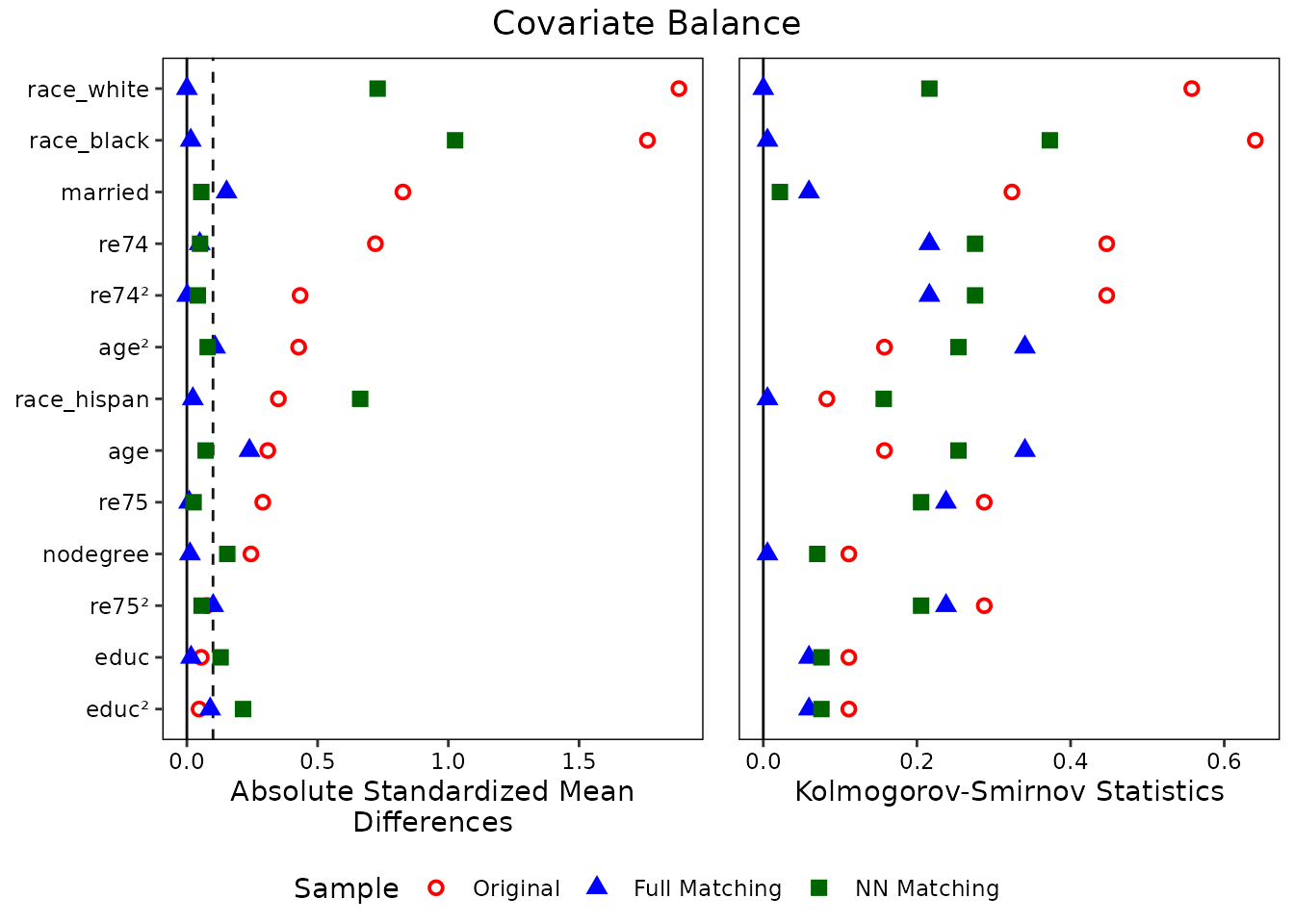
The love.plot() documentation explains what each of
these arguments do and the several other ones available. See
vignette("love.plot", package = "cobalt") for other
advanced customization of love.plot().
bal.plot()
bal.plot() displays distributional balance for a single
covariate, similar to plot.matchit(). Its default is to
display kernel density plots for continuous variables and bar graphs for
categorical variables. It can also display eCDF plots and histograms.
Below we demonstrate some of its uses:
#Density plot for continuous variables
bal.plot(m.out, var.name = "educ", which = "both")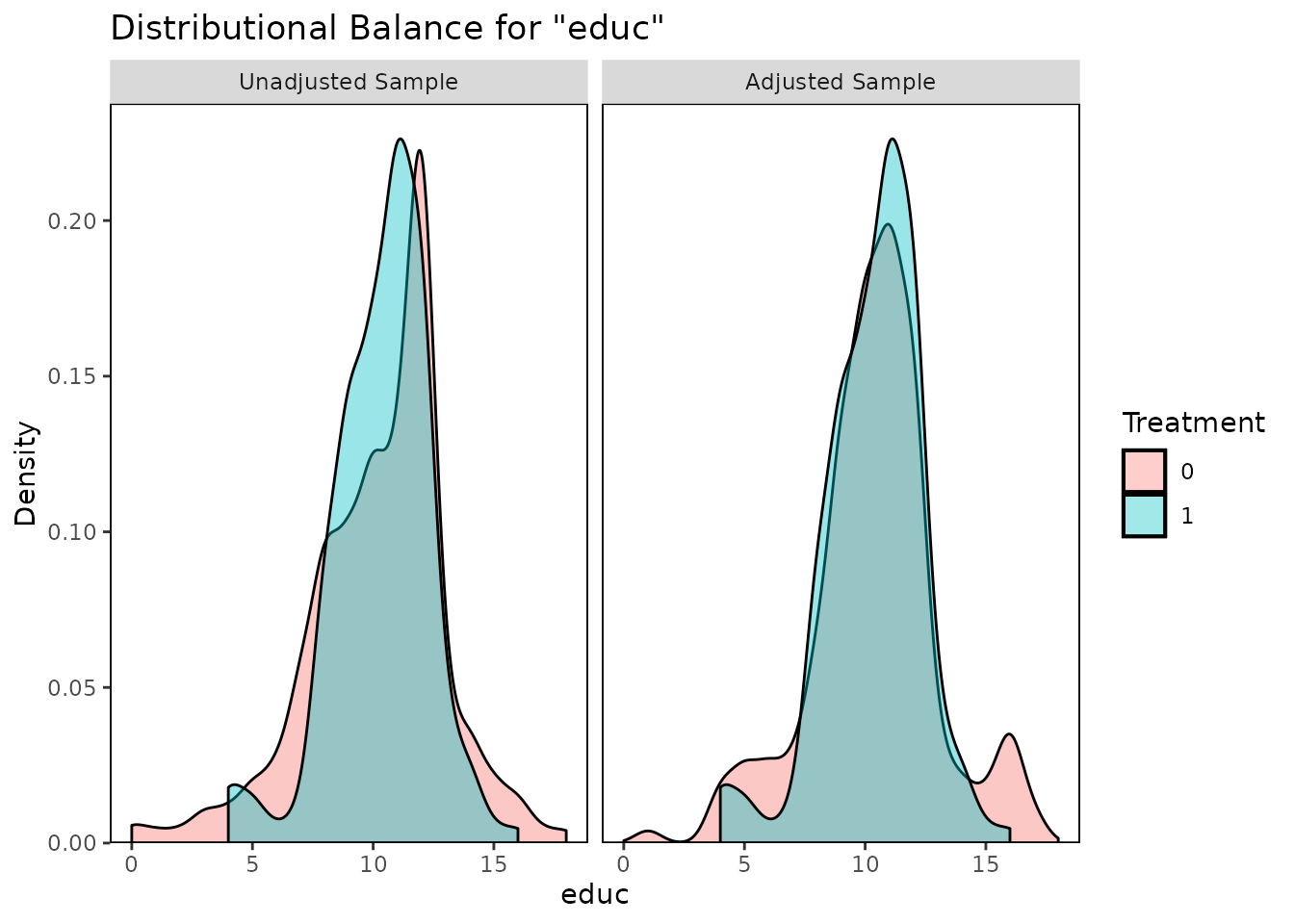
#Bar graph for categorical variables
bal.plot(m.out, var.name = "race", which = "both")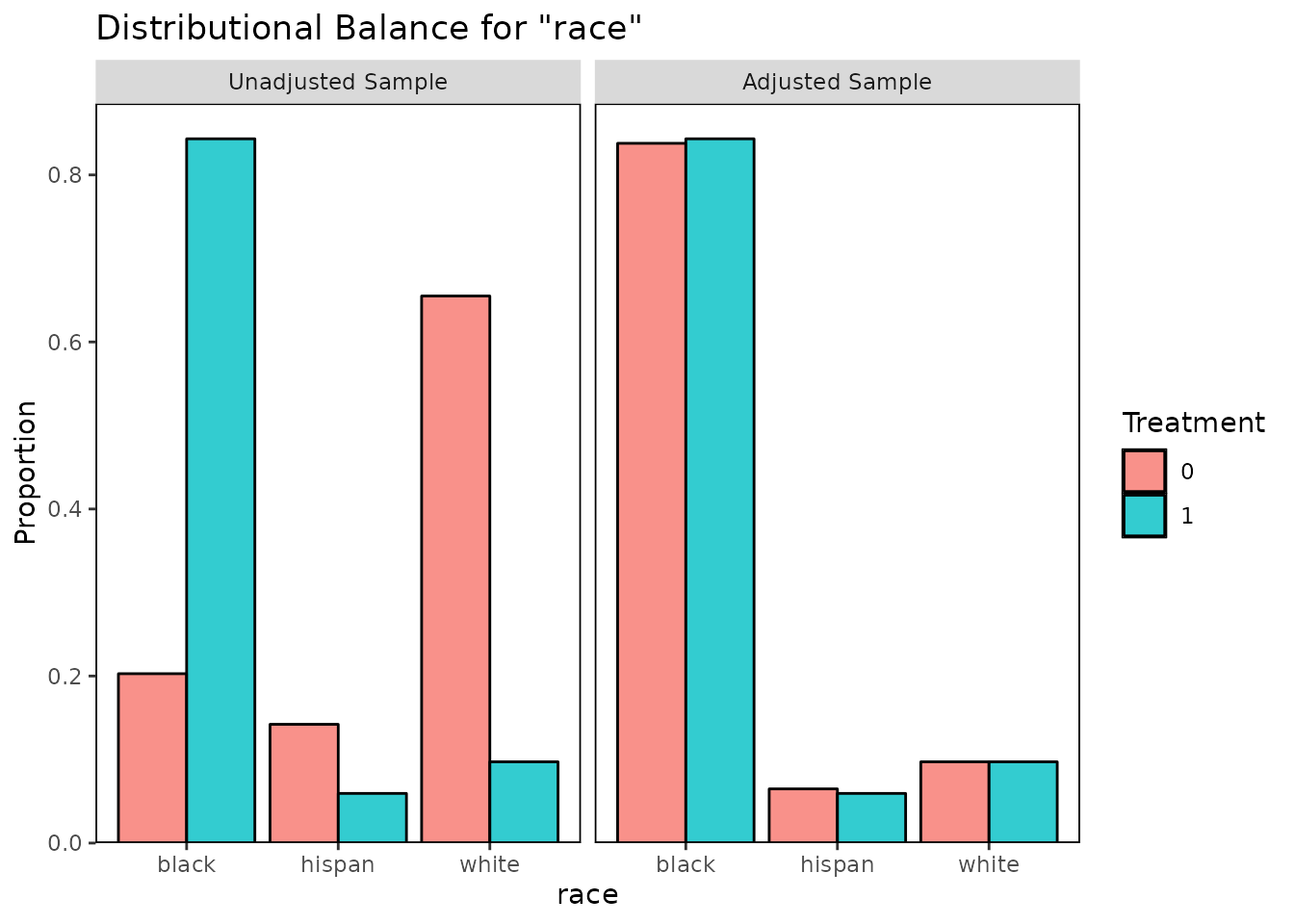
#Mirrored histogram
bal.plot(m.out, var.name = "distance", which = "both",
type = "histogram", mirror = TRUE)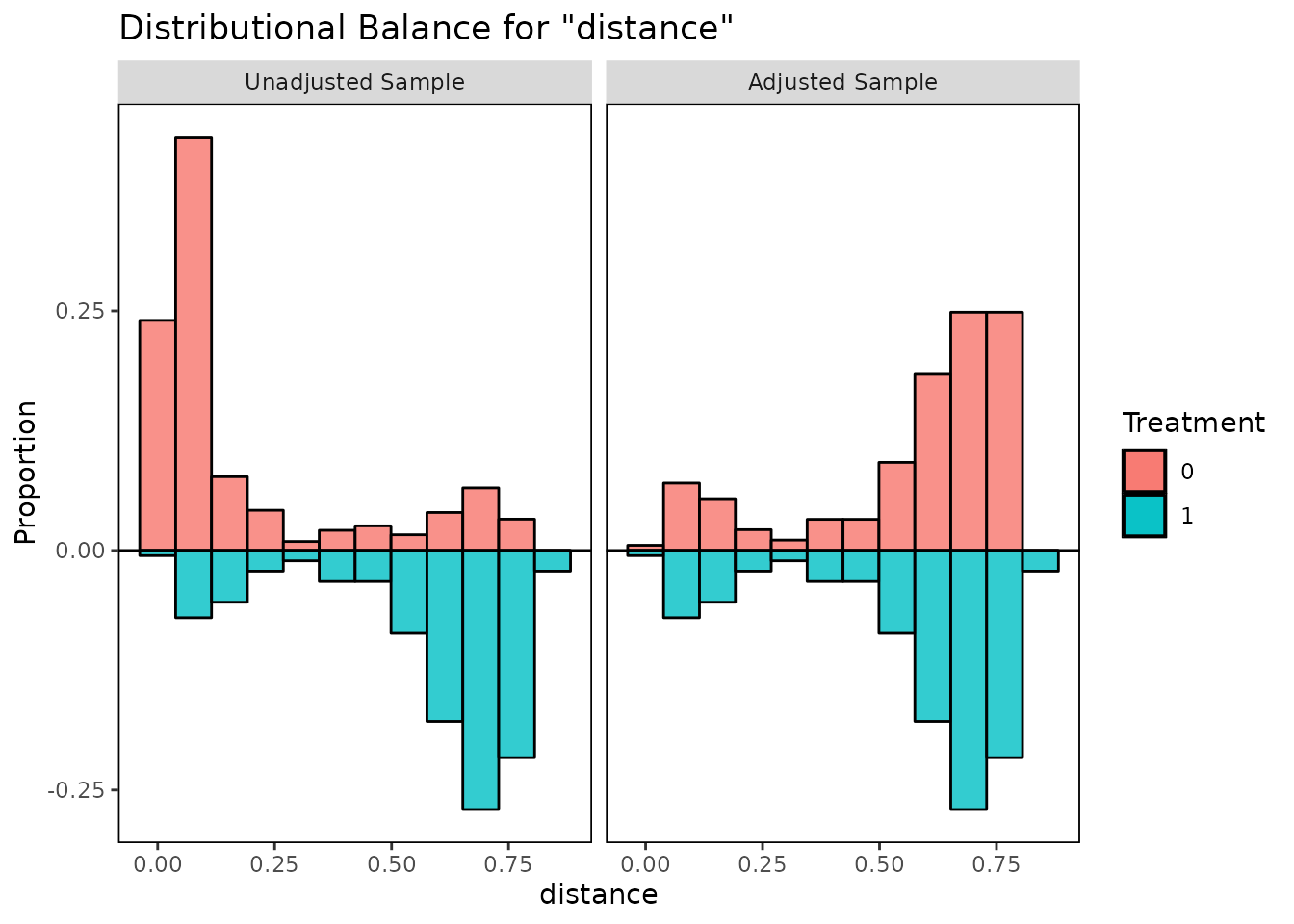
These plots help illuminate the specific ways in which the covariate
distributions differ between treatment groups, which can aid in
interpreting the balance statistics provided by bal.tab()
and summary.matchit().
Conclusion
The goal of matching is to achieve covariate balance, similarity
between the covariate distributions of the treated and control groups.
Balance should be assessed during the matching phase to find a matching
specification that works. Balance must also be reported in the write-up
of a matching analysis to demonstrate to readers that matching was
successful. MatchIt and cobalt each offer a
suite of functions to implement best practices in balance assessment and
reporting.